SFIHMM
high-speed C code for the estimation of Hidden Markov Models (finite state machines) on arbitrary time-series, for Viterbi Path Reconstruction, PCCA+ (Perron-Cluster Cluster Analysis), and for the generation of simulated data from HMMs. Based on the code used in "Conflict and Computation on Wikipedia: a Finite-State Machine Analysis of Editor Interactions”, where these methods were used to detect critical transitions in symbolic time series.Now includes glue code for parallel use of multiple cores.
This is a pre-release version of the code; feedback and requests for additional features solicited; please write sdedeo[at]andrew.cmu.edu
1. Installing
2. Getting started: estimating an HMM from data
3. How many hidden states?
4. Parallel SFIHMM
5. Viterbi Path Reconstruction
6. Module Identification — Critical Transition detection
7. Generating new sequences
8. Polishing solutions
9. Notes / Citing / Acknowledgements
10. Help and Questions From Users
Installing
SFIHMM works on Mac OS X and UNIX systems such as GNU/Linux. You’ll need the gsl libraries. On the Mac, you can load these in using tools such as macports (sudo port install gsl) or homebrew. You’ll know it’s working if you can run the command gsl-config. We know it works on both Ubuntu and Gentoo Linux, and CentOS, as well as Mac OS X; for Linux boxes you may need to install (via apt-get) gsl-bin, libgsl0-dev, and libgsl0ldb.Once you’ve installed gsl, download SFIHMM.zip, and uncompress by typing "unzip SFIHMM.zip". Type "cd SFIHMM" (i.e., go to the SFIHMM directory), and type "make". You should now be able to run ./hmm when inside the SFIHMM directory. You’re off and running! I’ve included a test file, test_data, so you can experiment with the different options available for the code.
Estimating a Hidden Markov Model (pre-specified number of states)
The most basic task: given a sequence, find the best-fitting Hidden Markov Model that describes it.ayerie:~simon$ ./hmm -f [N_iterations] [N_states] [name of data file]
Input
data file. An ASCII text file with two lines; I’ve given you a sample file, test_data, to play with, but you’ll want to use your own eventually!Line One: the total number of observations in the sequence.
Line Two: the sequence itself. The code reads in all the symbols, and orders them alphabetically (by ASCII order). Don’t use multi-letter symbols!—you may need to pre-process your data to create a proper input file to SFIHMM. For example, you can use all of the letters from A to Z, and the all of the lowercase letters from a to z, to get up to 52 different symbols. If a symbol doesn’t appear in the input stream, SFIHMM can’t know it exists, so (e.g.) if you have A, B, and D in the data, then SFIHMM will call A symbol one, B symbol two, and D symbol three. Beware!
Command line options
[N_iterations]: the number of times to run SFIHMM’s EM algorithm from random initial conditions. Landscapes are "rugged", meaning that there can be multiple solutions, some of which are sub-optimal. By increasing the number of times you run SFIHMM, you increase your chances of finding the optimal machine—at the cost of running the algorithm for longer, of course![N_states]: the number of states in the final output.
Example:
ayerie:~simon$ ./hmm -f 100 5 test_data
Fit a five-state machine; run the algorithm 100 times; return the highest-likelihood machine.
Output
What you want is sent to the file [name of file]_OUT_[N_states]states ; for the example above, that would be test_data_OUT_5states. It is an ASCII text file listing, in order:line 1: the log-likelihood
line 2: the number of states, N
line 3: the number of output symbols, M
starting at line 4: the state-to-state transition probabilities (the probability of going from state 0 to state 3, for example, can be found on line 4, column 4). There are N rows, one for each state.
starting at line 4+N: the symbol emission probabilities for each state (the probability of emitting symbol two in state zero, for example, can be found at line 4+N, column three). There are N rows, one for each state.
line 4+2N: the list of symbols in order (this tells you which symbol is state zero, state one, etc);
line 5+2N: the stationary distribution over the hidden states
line 6+2N: λ2, the second eigenvalue (see Equation 1 of the Wikipedia paper)
Estimating a Hidden Markov Model (without specifying the number of states)—recommended!
What happens when you don’t have a good idea how many states you should use? Too few, and you miss important patterns; too many, and you are in danger of overfitting to noise. The general problem of how to select the correct number of states is called "model selection". One model selection solution (far from the only one) is to use AIC (the "Akaike Information Criterion"). SFIHMM will do this for you—all you have to do is leave off the [N_states] option:ayerie:~simon$ ./hmm -f [N_iterations] [name of data file]
For example:
ayerie:~simon$ ./hmm -f 100 test_data
I’d recommend using the AIC model selection tool rather than pre-specifying the number of states. It is hard to get an intuition about the "right" number of states to use; tests in our paper suggest that AIC does very well in approximating the true number of states.
Parallel SFIHMM: fitting HMMs with multiple cores
SFIHMM now has a little ruby glue code to run the -f option in parallel. This is in test mode! It should be tidy and clean up after itself.ayerie:~simon$ ./parallel_glue.rb [number of cores] [number of iterations per core] [filename]
For example:
ayerie:~simon$ ./parallel_glue.rb 8 10 test_data
To state the obvious: you can either run on multiple cores with the same number of iterations (takes the same amount of time, but better fits); or with a smaller number of iterations (if the number of cores times the number of iterations is the same, the fit quality won’t decline, but it will run ncores times faster.)
If you want to use the parallel code on a specified range of state sizes, you can now run
ayerie:~simon$ ./parallel_glue.rb [number of cores] [number of iterations per core] [filename] [start N_states] [finish N_states]
For example:
ayerie:~simon$ ./parallel_glue.rb 8 10 test_data 7 10
will run the code looking a state sizes seven through ten (and selecting the best state size based on AIC), while
ayerie:~simon$ ./parallel_glue.rb 8 10 test_data 7 7
will just try the seven state case.
Viterbi path reconstruction
Say you’ve found a good model to describe your data. As the timeseries unfolds, what internal state is the system most likely to be in over time? Viterbi Path Reconstruction finds the most likely path through the system, given a set of data, and an accompanying model. Note that the data you feed it doesn’t have to be the same data you used to build the model! (But it can’t have any new symbols the original model didn’t see.) Use the -v ("Viterbi") option.ayerie:~simon$ ./hmm -v [name of data file] [name of HMM file]
For example:
ayerie:~simon$ ./hmm -v test_data test_data_OUT_4states
The output is a list of numbers, specifying which hidden state the system is most likely to be in at each step in the time-series.
Module identification and path reconstruction
Depending on your system, the relaxation time may be very long, and the system may be trapped in a subspace of the available states. Using the methods described in the Wikipedia paper (see Sec. 1.4), we can use the second eigenvector to find the two main system modules. Using the -m ("Module") option, SFIHMM will identify these modules; it will name one H ("Higher probability of emitting symbol 1") and the other L, and output the path the system takes between themayerie:~simon$ ./hmm -m [name of data file] [name of HMM file]
For example:
ayerie:~simon$ ./hmm -m test_data test_data_OUT_4states
The output is a list of letters (H or L), specifying which module the system is most likely to be in at each step in the time-series. It will also tell you the state membership in the two modules, the probability of emitting the first symbol conditional upon being within the module (using the stationary distribution), and the empirical trapping times in the two states.
Generating new sequences
Say you’ve found a good model to describe your data, and you’d like to generate more of it. SFIHMM is happy to help; with the -g ("generate") option, tell SFIHMM the number of letters you’d like, and feed it the HMM you desire.ayerie:~simon$ ./hmm -g [N_letters] [name of HMM file]
For example:
ayerie:~simon$ ./hmm -g 100 test_data_OUT_4states
Polishing solutions
The standard SFIHMM algorithm terminates when it can not improve the solution beyond a tolerance of 10-3 in log-likelihood. After finding a solution, however, you can polish it using the -p option:ayerie:~simon$ ./hmm -p [name of data file] [name of HMM file] [tolerance]
For example:
ayerie:~simon$ ./hmm -p test_data test_data_OUT_4states 1e-6
Notes
I’ve written SFIHMM with the goal of maximal simplicity. For example, you don’t specify the symbol space; SFIHMM figures it out from the data itself. And its output HMM descriptions are also valid file formats for input.Citation
SFIHMM was written for the calculations first described and presented in this paper, including the use of Viterbi path reconstruction, and λ2 / trapping time characterization.BibTeX
@Article{dedeo16,AUTHOR = {DeDeo, Simon},
TITLE = {Conflict and Computation on {W}ikipedia: A Finite-State Machine Analysis of Editor Interactions},
JOURNAL = {Future Internet},
VOLUME = {8},
YEAR = {2016},
NUMBER = {3},
PAGES = {31},
URL = {http://www.mdpi.com/1999-5903/8/3/31},
ISSN = {1999-5903},
DOI = {10.3390/fi8030031}
}
Acknowledgements
The development of SFIHMM was supported in part by the National Science Foundation under Grant No. EF-1137929, "the small-number limit of biological information-processing", and by the Santa Fe Institute through an Omidyar Fellowship award to Simon DeDeo. Any opinions, findings and conclusions or recommendations expressed by SFIHMM are those of SFIHMM and do not necessarily reflect the views of the National Science Foundation (NSF) or any other funders. I thank Anne Sallaska, David Slater, and Haven Liu (MITRE Corp) for beta-testing this code.Help and Questions from Users
Hi Simon,[…] As part of my thesis, I want to detect times of conflict within Wikipedia articles. I read your recent article "Conflict and Computation on Wikipedia: A Finite-State Machine Analysis of Editor Interactions", and found the general method you're employing to be remarkable, and maybe of great use for me. Now, I'm not a programmer, and my math skills aren't good enough as to understand the formulas you used, but the fact that this method can distinguish conflict times from "normal" times is precisely what I need. What's even better is that you supplied the data and code for analysing the raw data. I downloaded both the SFIHMM program and the dataset of your article. Thank you for that!
Since I'm no programmer (and I've yet to install a UNIX OS), as I mentioned, I wanted to ask some questions:
-Does the SFIHMM program able to create the .tsv edit files from .xml history files of Wikipedia articles? (or for that matter, from any other raw file)
No—you’ll have to create these yourself. Take a look at the “test_data” file in the ZIP that comes with SFIHMM. Then you might also look at http://tuvalu.santafe.edu/~simon/styled-9/styled-12/ which contains some example TSV files, and the derived input files for the SFIHMM code.
-Is it possible to know from one of the files created by SFIHMM when a transition from the state of cooperation to conflict (and vice versa) occurs?
Yes: SFIHMM will find these for you. Take a look at the “Module Identification and path reconstruction” section above. The H and L symbols are the derived high and low conflict states, respectively.
-I couldn't really make what the number on the [Filename]_hmm_[X]states files mean. Could you explain very shortly? Or even refer me to some explanation I might have missed.
The number, X, is the number of hidden states in the model. In the paper, for example, you can see that the George_W._Bush example has twelve hidden states. If you don’t want to specify the number of states in advance, take a look at Estimating a Hidden Markov Model (without specifying the number of states) above for a way to have the machine itself infer the optimal number.
I really hope all these questions aren't a burden on you.
Thank you very much in advance,
Ben
Not at all—good luck.
<<<<<
Thank you very much for your reply.
I have looked at some .tsv files and it seemed to me that the tagging of each edit as C or R was done automatically with some code. From what I saw in in Appendix S3 of your other article (Collective Phenomena and Non Finite State Computation in a Human Social System) you use the comments of each edit and identical SHA1 to identify reverts. Did you do that manually?
Well, not quite manually!—I wrote some code. It’s pretty simple: define a “revert” as an edit that takes a page back to a SHA1 Hash you’ve already seen in the edits so far. (That’s not quite perfect, because there are a few edge cases, e.g., someone makes an edit and reverts themselves, but these are pretty rare.) The SHA1s can be retrieved using the API; in the tsvs I believe they’re included.
About my third question, I think I did not explain myself well enough, so I added a screenshot (Lebanon war, 6 states) of the numbers I don't understand (I only understand that there is a column for each state):
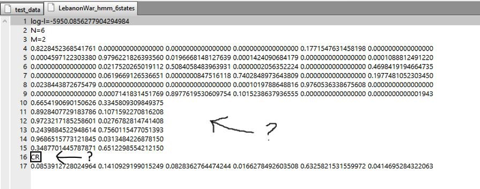
:) OK, so in your image there, lines 10 through 15 are the probabilities that each state emits either C or R. For example, the probability that the system emits an R when in state three is 2.7%. The “CR” on line 16 tells you the order of the symbols (i.e., because C is first, you know that the first entry on line 17 refers to the probability of emitting C.)
Again, thank you very much!
Hope this helps!
>>>
Hello Simon,
I'm trying to use SFIHMM for my experiments on large traces (300K of length) from a hexaxopter. I tried using the ruby glue to parallelize the training, and it seemed to work up to 10 states (I run it without specifying the number of states), but then I got this error:
./parallel_glue.rb:35:in ``': Cannot allocate memory - ls ../Traces/Hexacopter/full_while_kernel_calls.csv_MULTI*_OUT_10states 2 /dev/null (Errno::ENOMEM)
from ./parallel_glue.rb:35:in `block in main'
from ./parallel_glue.rb:28:in `upto'
from ./parallel_glue.rb:28:in `main'
I tried to find the root-cause of the problem, but I couldn't make sense of the output quite well. So I run SFIHMM on 10 cores each with 100 iterations (./parallel_glue.rb 10 100 ../Traces/Hexicopter/full_while_kernel_calls.csv), and I see several output files for each state (for example full_while_kernel_calls.csv_MULTI0_OUT_10states). And each output has its own fit (though with very close log-likelihood). Now I was expecting that, say for 10 states, I'll get a final single output that has merged the fits from all iterations, but it seems that SFIHMM produces several separate outputs from each core, and I'm supposed to merge them together, is that right?
If that's the case, why the output MULTI*_OUT_nstates does not exist for all states? For example for 2 states I only have full_while_kernel_calls.csv_MULTI0_OUT_2states, but for 3 states I have full_while_kernel_calls.csv_MULTI1_OUT_3states and full_while_kernel_calls.csv_MULTI5_OUT_3states. Is that because, for 2 states, the iterations from one core were enough to find an fit?
I general I want to make sense of the output of the ruby glue.
Thanks in advance for your help Simon!
Reza
<<<<
Hello Reza —
Thank you for writing, and happy to hear that the code is getting a workout. So, it certainly looks like you’re running out of memory when you first do it. Have you tried running just on two cores, say? This should give you five times more memory, and would be a good way to start figuring out what’s going on and learn the code a bit.
If the program crashes in the middle of finding solutions, you’ll find that there are lots of MULTI*_OUT files hanging around. This is because the code didn’t do all the iterations; it will only pick the best solution once all the cores have done their work. In the middle, it will keep all the outputs. You should try to solve the problem by using fewer cores, so that you don’t run out of memory.
If the code completes correctly, there should be only one HMM file, which will have a name like (in your case) full_while_kernel_calls.csv_OUT_12states, without the MULTI, if the twelve state model is the best AIC fit.
My suggestion is to first run the demo “test_data” file, i.e., go to the SFIHMM directory and run "./parallel_glue.rb 10 100 test_data”. Then, try your case, but on only one core, "./parallel_glue.rb 1 100 ../Traces/Hexicopter/full_while_kernel_calls.csv”
Let me know if this works,
Simon
>>>>
Also is there a way to run the parallel version with the number of states specified?
Sure! I’ve just added this option—you can now specify the range. If you want to just do five states, try
./parallel_glue.rb 1 100 test_data 5 5
if you want to do a range of states try
./parallel_glue.rb 1 100 test_data 5 7

